The ao package implements alternating optimization in R. This vignette demonstrates how to use the package and describes the available customization options.
What is alternating optimization?
Alternating optimization (AO) is an iterative process for optimizing a multivariate function by breaking it down into simpler sub-problems. It involves optimizing over one block of function parameters while keeping the others fixed, and then alternating this process among the parameter blocks. AO is particularly useful when the sub-problems are easier to solve than the original joint optimization problem, or when there is a natural partitioning of the parameters. See Bezdek and Hathaway (2002), Hu and Hathaway (2002), and Bezdek and Hathaway (2003) for more details.
Consider a real-valued objective function where and are two blocks of function parameters, namely a partition of the parameters. The AO process can be described as follows:
Initialization: Start with initial guesses and .
-
Iterative Steps: For
- Step 1: Fix and solve the sub-problem
- Step 2: Fix and solve the sub-problem
Convergence: Repeat the iterative steps until a convergence criterion is met, such as when the change in the objective function or the parameters falls below a specified threshold, or when a pre-defined iteration limit is reached.
The AO process can be
viewed as a generalization of joint optimization, where the parameter partition is trivial, consisting of the entire parameter vector as a single block,
also used for maximization problems by simply replacing by above,
generalized to more than two parameter blocks, i.e., for , the process involves cycling through each parameter block and solving the corresponding sub-problems iteratively (the parameter blocks do not necessarily have to be disjoint),
randomized by changing the parameter partition randomly after each iteration, which can further improve the convergence rate and help avoid getting trapped in local optima (Chib and Ramamurthy 2010),
run in multiple processes for different initial values, parameter partitions, and/or base optimizers.
How to use the package?
The ao package provides a single user-level function,
ao(), which serves as a general interface for performing
various variants of AO.
The function call
The ao() function call with the default arguments looks
as follows:
ao(
f,
initial,
target = NULL,
npar = NULL,
gradient = NULL,
hessian = NULL,
...,
partition = "sequential",
new_block_probability = 0.3,
minimum_block_number = 1,
minimize = TRUE,
lower = NULL,
upper = NULL,
iteration_limit = Inf,
seconds_limit = Inf,
tolerance_value = 1e-6,
tolerance_parameter = 1e-6,
tolerance_parameter_norm = function(x, y) sqrt(sum((x - y)^2)),
tolerance_history = 1,
base_optimizer = Optimizer$new("stats::optim", method = "L-BFGS-B"),
verbose = FALSE,
hide_warnings = TRUE,
add_details = TRUE
)The arguments have the following meaning:
f: The objective function to be optimized. By default,fis optimized over its first argument. If optimization should target a different argument or multiple arguments, usenparandtarget, see below. Additional arguments forfcan be passed via the...argument as usual.initial: Initial values for the parameters used in the AO process.gradientandhessian: Optional arguments to specify the analytical gradient and/or Hessian off.-
partition: Specifies how parameters are partitioned for optimization. Can be one of the following:"sequential": Optimizes each parameter block sequentially. This is similar to coordinate descent."random": Randomly partitions parameters in each iteration."none": No partitioning; equivalent to joint optimization.Custom partition can be defined using a list of vectors of parameter indices, see below.
new_block_probabilityandminimum_block_numberare only relevant ifpartition = "random". In this case, the former controls the probability for creating a new block when building a random parameter partition, and the latter defines the minimum number of parameter blocks in the partition.minimize: Set toTRUEfor minimization (default), orFALSEfor maximization.lowerandupper: Lower and upper limits for constrained optimization.iteration_limitis the maximum number of AO iterations before termination, whileseconds_limitis the time limit in seconds.tolerance_valueandtolerance_parameter(in combination withtolerance_parameter_norm) specify two other stopping criteria, namely when the difference between the current function value or the current parameter vector and the one beforetolerance_historyiterations, respectively, becomes smaller than these thresholds.base_optimizer: Numerical optimizer used for solving sub-problems, see below.Set
verbosetoTRUEto print status messages, andhide_warningstoFALSEto show warning messages during the AO process.add_details = TRUEadds additional details about the AO process to the output.
A simple first example
The following is an implementation of the Himmelblau’s function
himmelblau <- function(x) (x[1]^2 + x[2] - 11)^2 + (x[1] + x[2]^2 - 7)^2This function has four identical local minima, for example in and :
himmelblau(c(3, 2))
#> [1] 0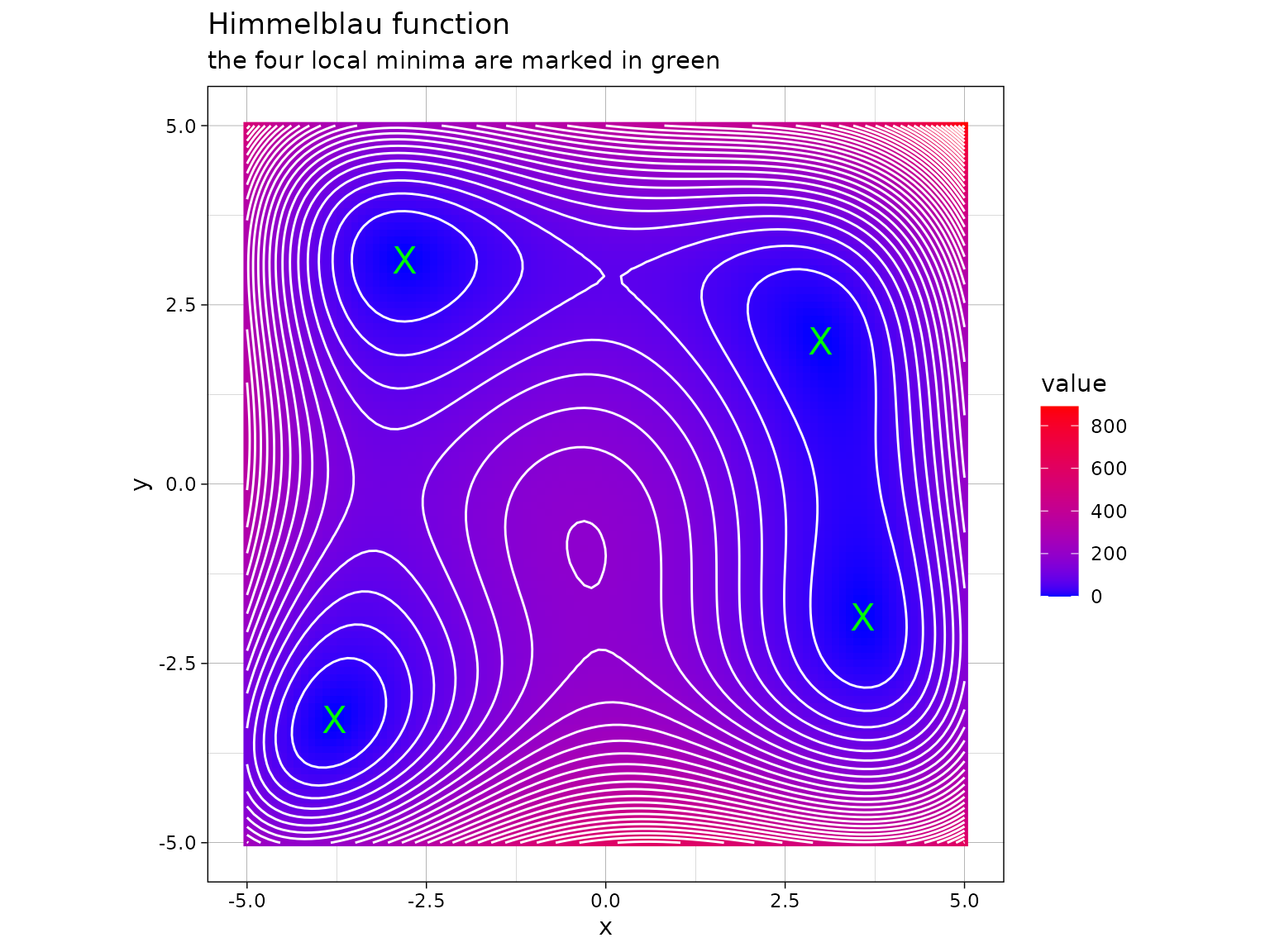
Minimizing Himmelblau’s function through alternating minimization over and with initial values can be accomplished as follows:
ao(f = himmelblau, initial = c(0, 0))
#> $estimate
#> [1] 3.584428 -1.848126
#>
#> $value
#> [1] 9.606386e-12
#>
#> $details
#> iteration value p1 p2 b1 b2 seconds
#> 1 0 1.700000e+02 0.000000 0.000000 0 0 0.000000000
#> 2 1 1.327270e+01 3.395691 0.000000 1 0 0.046252251
#> 3 1 1.743664e+00 3.395691 -1.803183 0 1 0.010555267
#> 4 2 2.847290e-02 3.581412 -1.803183 1 0 0.007988214
#> 5 2 4.687468e-04 3.581412 -1.847412 0 1 0.006888866
#> 6 3 7.368057e-06 3.584381 -1.847412 1 0 0.005846977
#> 7 3 1.164202e-07 3.584381 -1.848115 0 1 0.054032564
#> 8 4 1.893311e-09 3.584427 -1.848115 1 0 0.004757404
#> 9 4 9.153860e-11 3.584427 -1.848124 0 1 0.003518105
#> 10 5 6.347425e-11 3.584428 -1.848124 1 0 0.003552437
#> 11 5 9.606386e-12 3.584428 -1.848126 0 1 0.003607750
#>
#> $seconds
#> [1] 0.1469998
#>
#> $stopping_reason
#> [1] "change in function value between 1 iteration is < 1e-06"Here, we see the output of the AO process, which is a
list that contains the following elements:
estimateis the parameter vector at termination.valueis the function value at termination.detailsis adata.framewith full information about the process: For each iteration (columniteration) it contains the function value (columnvalue), parameter values (columns starting withpfollowed by the parameter index), the active parameter block (columns starting withbfollowed by the parameter index, where1stands for a parameter contained in the active parameter block and0if not), and computation times in seconds (columnseconds).secondsis the overall computation time in seconds.stopping_reasonis a message why the process has terminated.
Using the analytical gradient
For the Himmelblau’s function, it is straightforward to define the analytical gradient as follows:
gradient <- function(x) {
c(
4 * x[1] * (x[1]^2 + x[2] - 11) + 2 * (x[1] + x[2]^2 - 7),
2 * (x[1]^2 + x[2] - 11) + 4 * x[2] * (x[1] + x[2]^2 - 7)
)
}The gradient function is used by ao() if provided via
the gradient argument as follows:
ao(f = himmelblau, initial = c(0, 0), gradient = gradient, add_details = FALSE)
#> $estimate
#> [1] 3.584428 -1.848126
#>
#> $value
#> [1] 2.659691e-12
#>
#> $seconds
#> [1] 0.06115723
#>
#> $stopping_reason
#> [1] "change in function value between 1 iteration is < 1e-06"In scenarios involving higher dimensions, utilizing the analytical gradient can notably improve both the speed and stability of the process. The analytical Hessian can be utilized analogously.
Random parameter partitions
Another version of the AO process involves using a new, random
partition of the parameters in every iteration. This approach can
enhance the convergence rate and prevent being stuck in local optima. It
is activated by setting partition = "random". The
randomness can be adjusted using two parameters:
new_block_probabilitydetermines the probability for creating a new block when building a new partition. Its value ranges from0(no blocks are created) to1(each parameter is a single block).minimum_block_numbersets the minimum number of parameter blocks for random partitions. Here, it is configured as2to avoid generating trivial partitions.
The random partitions are build as follows:1
process <- ao:::Process$new(
npar = 10,
partition = "random",
new_block_probability = 0.5,
minimum_block_number = 2
)
process$get_partition()
#> [[1]]
#> [1] 5
#>
#> [[2]]
#> [1] 1 6 9
#>
#> [[3]]
#> [1] 10
#>
#> [[4]]
#> [1] 7
#>
#> [[5]]
#> [1] 4 8
#>
#> [[6]]
#> [1] 2 3
process$get_partition()
#> [[1]]
#> [1] 1 7 8
#>
#> [[2]]
#> [1] 6 10
#>
#> [[3]]
#> [1] 3 4
#>
#> [[4]]
#> [1] 2
#>
#> [[5]]
#> [1] 9
#>
#> [[6]]
#> [1] 5As an example of AO with random partitions, consider fitting a two-class Gaussian mixture model via maximizing the model’s log-likelihood function
where the sum goes over all observations , and denote the normal density for the first and second cluster, respectively, and is the mixing proportion. The parameter vector to be estimated is . As there exists no closed-form solution for the maximum likelihood estimator , we apply numerical optimization to find the function optimum. The model is fitted to the following data:2
The following function calculates the log-likelihood value given the
parameter vector theta and the observation vector
data:3
normal_mixture_llk <- function(theta, data) {
mu <- theta[1:2]
sd <- exp(theta[3:4])
lambda <- plogis(theta[5])
c1 <- lambda * dnorm(data, mu[1], sd[1])
c2 <- (1 - lambda) * dnorm(data, mu[2], sd[2])
sum(log(c1 + c2))
}The ao() call for performing alternating
maximization with random partitions looks as follows, where we
simplified the output for brevity:
out <- ao(
f = normal_mixture_llk,
initial = runif(5),
data = datasets::faithful$eruptions,
partition = "random",
minimize = FALSE
)
round(out$details, 2)
#> iteration value p1 p2 p3 p4 p5 b1 b2 b3 b4 b5 seconds
#> 1 0 -713.98 0.94 0.79 0.97 0.35 0.50 0 0 0 0 0 0.00
#> 2 1 -541.18 0.94 3.81 0.97 0.35 0.50 0 1 0 0 0 0.01
#> 3 1 -512.65 0.94 3.81 0.66 -0.30 0.50 0 0 1 1 0 0.02
#> 4 1 -447.85 3.08 3.81 0.66 -0.30 0.50 1 0 0 0 0 0.01
#> 5 1 -445.29 3.08 3.81 0.66 -0.30 -0.04 0 0 0 0 1 0.01
#> 6 2 -277.37 2.02 4.27 -1.55 -0.83 -0.55 1 1 1 1 1 0.08
#> 7 3 -276.36 2.02 4.27 -1.45 -0.83 -0.63 1 1 1 1 1 0.07
#> 8 4 -276.36 2.02 4.27 -1.45 -0.83 -0.63 0 1 0 0 0 0.00
#> 9 4 -276.36 2.02 4.27 -1.45 -0.83 -0.63 0 0 1 0 1 0.01
#> 10 4 -276.36 2.02 4.27 -1.45 -0.83 -0.63 1 0 0 1 0 0.01
#> 11 5 -276.36 2.02 4.27 -1.45 -0.83 -0.63 1 0 0 1 1 0.01
#> 12 5 -276.36 2.02 4.27 -1.45 -0.83 -0.63 0 0 1 0 0 0.00
#> 13 5 -276.36 2.02 4.27 -1.45 -0.83 -0.63 0 1 0 0 0 0.00
#> 14 6 -276.36 2.02 4.27 -1.45 -0.83 -0.63 1 1 0 1 0 0.01
#> 15 6 -276.36 2.02 4.27 -1.45 -0.83 -0.63 0 0 0 0 1 0.00
#> 16 6 -276.36 2.02 4.27 -1.45 -0.83 -0.63 0 0 1 0 0 0.00More flexibility
The ao package offers some flexibility for performing AO.4
Generalized objective functions
Optimizers in R generally require that the objective function has a
single target argument which must be in the first position, but
ao allows for optimization over an argument other than
the first, or more than one argument. For example, say, the
normal_mixture_llk function above has the following form
and is supposed to be optimized over the parameters mu,
lsd, and llambda:
normal_mixture_llk <- function(data, mu, lsd, llambda) {
sd <- exp(lsd)
lambda <- plogis(llambda)
c1 <- lambda * dnorm(data, mu[1], sd[1])
c2 <- (1 - lambda) * dnorm(data, mu[2], sd[2])
sum(log(c1 + c2))
}In ao(), this scenario can be specified by setting
target = c("mu", "lsd", "llambda")(the names of the target arguments)and
npar = c(2, 2, 1)(the lengths of the target arguments):
Parameter bounds
Instead of using parameter transformations in the
normal_mixture_llk() function above, parameter bounds can
be specified via the arguments lower and
upper, where both can either be a single number (a common
bound for all parameters) or a vector of specific bounds per parameter.
Therefore, an more straightforward implementation of the mixture example
would be:
normal_mixture_llk <- function(mu, sd, lambda, data) {
c1 <- lambda * dnorm(data, mu[1], sd[1])
c2 <- (1 - lambda) * dnorm(data, mu[2], sd[2])
sum(log(c1 + c2))
}
ao(
f = normal_mixture_llk,
initial = runif(5),
target = c("mu", "sd", "lambda"),
npar = c(2, 2, 1),
data = datasets::faithful$eruptions,
partition = "random",
minimize = FALSE,
lower = c(-Inf, -Inf, 0, 0, 0),
upper = c(Inf, Inf, Inf, Inf, 1)
)Custom parameter partition
Say the parameters of the Gaussian mixture model are supposed to be grouped by type:
In ao(), custom parameter partitions can be specified by
setting partition = list(1:2, 3:4, 5), i.e. by defining a
list where each element corresponds to a parameter block,
containing a vector of parameter indices. Parameter indices can be
members of any number of blocks.
Stopping criteria
Currently, four different stopping criteria for the AO process are implemented:
a predefined iteration limit is exceeded (via the
iteration_limitargument)a predefined time limit is exceeded (via the
seconds_limitargument)the absolute change in the function value in comparison to the last iteration falls below a predefined threshold (via the
tolerance_valueargument)the change in parameters in comparison to the last iteration falls below a predefined threshold (via the
tolerance_parameterargument, where the parameter distance is computed via the norm specified astolerance_parameter_norm)
Any number of stopping criteria can be activated or deactivated5, and the final output contains information about the criterium that caused termination.
Base optimizer
By default, the L-BFGS-B algorithm (Byrd et al. 1995) implemented in
stats::optim is used for solving the sub-problems
numerically. However, any other optimizer can be selected by specifying
the base_optimizer argument. Such an optimizer must be
defined through the framework provided by the optimizeR
package, see its
documentation for details. For example, the stats::nlm
optimizer can be selected by setting
base_optimizer = Optimizer$new("stats::nlm").
Multiple processes
AO can suffer from local optima. To increase the likelihood of reaching the global optimum, users can specify
multiple starting parameters,
multiple parameter partitions,
multiple base optimizers.
Use the initial, partition, and/or
base_optimizer arguments to provide a list of
possible values for each parameter. Each combination of initial values,
parameter partitions, and base optimizers will create a separate AO
process:
normal_mixture_llk <- function(mu, sd, lambda, data) {
c1 <- lambda * dnorm(data, mu[1], sd[1])
c2 <- (1 - lambda) * dnorm(data, mu[2], sd[2])
sum(log(c1 + c2))
}
out <- ao(
f = normal_mixture_llk,
initial = list(runif(5), runif(5)),
target = c("mu", "sd", "lambda"),
npar = c(2, 2, 1),
data = datasets::faithful$eruptions,
partition = list("random", "random", "random"),
minimize = FALSE,
lower = c(-Inf, -Inf, 0, 0, 0),
upper = c(Inf, Inf, Inf, Inf, 1)
)
names(out)
#> [1] "estimate" "estimates" "estimate_split" "value"
#> [5] "values" "details" "seconds" "seconds_each"
#> [9] "stopping_reason" "stopping_reasons" "processes"
out$values
#> [[1]]
#> [1] -421.417
#>
#> [[2]]
#> [1] -421.417
#>
#> [[3]]
#> [1] -585.2514
#>
#> [[4]]
#> [1] -276.36
#>
#> [[5]]
#> [1] -395.9705
#>
#> [[6]]
#> [1] -421.417In the case of multiple processes, the output provides information for the best process (with respect to the function value) as well as information on every single process.
By default, processes run sequentially. However, since they are
independent of each other, they can be parallelized. For parallel
computation, ao supports the {future}
framework. For example, run the following before the
ao() call:
future::plan(future::multisession, workers = 4)When using multiple processes, setting verbose = TRUE to
print tracing details during AO is not supported. However, progress of
processes can still be tracked using the {progressr}
framework. For example, run the following before the
ao() call:
progressr::handlers(global = TRUE)
progressr::handlers(
progressr::handler_progress(":percent :eta :message")
)